2023.12.08
News
A single-cell array can be achieved by “just a drop on a chip” - Hydrodynamic cell-trapping system in an open microfluidics architecture -

Abstract
Professor Hiroaki Suzuki of the Faculty of Science and Engineering, Chuo University, and collaborative researchers of the Graduate School of Science and Engineering, Tomoki Murakami (graduated in 2022), Hiroto Teratani, and Mamiko Tsugane, and the research group of Aeternus Co, Ltd and Caravell Co., Ltd have developed single-cell trapping in open microfluidics*1. The difference from the traditional closed microfluidics architecture is that trapped cells are exposed to the surface of the chip in the open microfluidics architecture, allowing an easy selection and retrieval of a specific cell using a versatile robotic picking device.
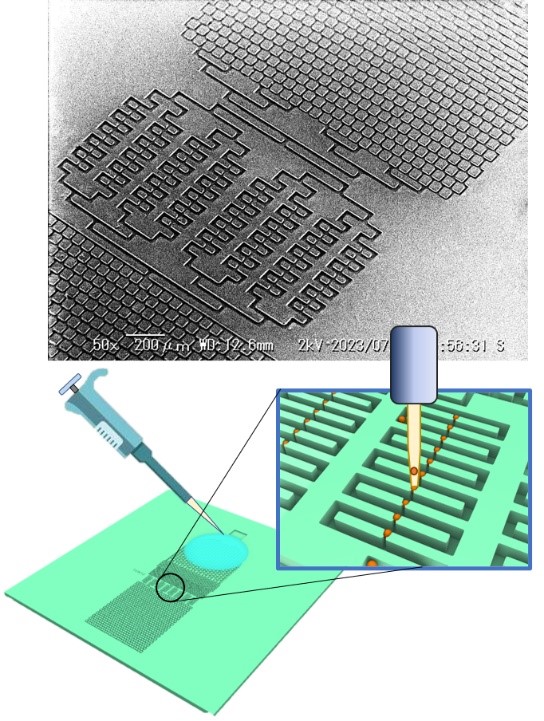
Scanning electron micrograph (upper panel) and a conceptual diagram of its usage (lower panerl) of the single-cell trapping device in an open microfluidics architecture.
Over the past decade, single-cell analysis and manipulation*2 technologies, in which single cells are examined and/or highly functional cells are selected, are developing rapidly. Among various existing technologies, an approach to select specific cells after microscope observation is orthodox but allows concrete and versatile screening. However, it has been difficult to process a large number of cells in an easy manner and in a short time.
In this study, we have solved the aforementioned issue by employing open microfluidics for trapping and arraying of single-cells. We established the microfluidics control method which leverages a spontaneous capillary flow (SCF) within narrow and shallow channels embedded on the chip surface, and designed the channel to trap cells on particular positions of the chip in a short time. Additionally we built a system to select and pick specific cells. This technology was realized by developing a precise hydrophilic treatment method applied only inside microchannels with the size of cells.
Thanks to this technology, we expect that screening technology*3 that selects and acquires highly functional cells will become available in various facilities, and will contribute to the realization of SDGs through bio-manufacturing.
This accomplishment was posted online on the International Academic Journal iScience as of October 24th, 2023 (EST).
************************************************************
【Researcher】
Hiroaki SUZUKI, Professor, Faculty of Science and Engineering, Chuo University (Department of Precision Mechanics)
【Article Information】
Publisher/Journal:iScience, Cell Press
Title: Single-Cell Trapping and Retrieval in Open Microfluidics
Research Content
1. Background
Over the past decade, single-cell analysis and manipulation have been developing at an accelerating pace. Leveraging single-cell analysis and manipulation, we can examine single cell's features and select highly functional cells. Among various existing technologies, the method to examine cells using a microscope and select them is orthodox but offer a robust and versatile screening platform. However, in order to identify and select/suction cells which are randomly distributed in a dish, a high-spec imaging apparatus and highly precise control of pipette positioning are inevitable. Futhremore, these processes tend to be slow. On the other hand, microfluidics technologies for single-cell trapping into arrays have been under intensive research, and this line of technology is expected to improve the efficiency of single-cell identification and observation. However, existing microfluidic devices employ the closed microchannels so it is impossible to retrieve cells from outside after cell trapping. Furthermore, closed systems require an external pump to drive the flow, which causes increase of the system size.
2. Research Contents and Accomplishments
The research group has solved the aforementioned issue by applying open microfluidics to single-cell trapping and arraying. We established the microfluidics control method which leverages a spontaneous capillary flow (SCF) within narrow and shallow channels embedded on the chip surface, and designed the channel to trap cells on particular positions of the chip in a short time. The size of conventional open microfluidics has been around 0.1~1mm, and single-cell trapping of 10 micrometers (0.01mm) has not been realized. The research group has developed a precise hydrophilic treatment method applied just inside the microchannels with the size close to cells and realized single-cell trapping in open microfluidics.
Specifically, we first formed the microscale trapping channel on the silicone rubber surface through the molding process (Figure 1). Then, we covered another silicone rubber plate on top of the channel and introduced the aqueous solution containing a hydrophilic agent. After a while, we removed the aqueous solution by suction and took off the lid to dry. Then, we can make only inside the microchannel hydrophilic engraved on the hydrophobic silicone rubber. As a result, spontaneous capillary flow occurs with just a drop of liquid on the edge of the channel. By designing microfluidic channel into the shape enabling single-cell/particle trapping, those particles or cells contained in the liquid are convected spontaneously inside the channel and trapped at specific positions (Figure 2). This procedure enables us to arrange cells into an array easily without a pump, and also thanks to its open surface, we can aspirate desirable cells using a glass capillary or pipette.
The research group has constructed the cell-picking system by combining the chip with a tabletop robot which suctions a particular cell or cell clusters using a glass pipette after arraying cells in precise positions on the chip (Figure 3).
In this article, we reproduced the design of two cell-trapping microfluidics (Tan et al., PNAS 2012 and Chung et al., Appl. Phys. Lett., 2011) established in the conventional closed microfluidics into the open microfluidics architecture in order to show versatility of this approach, and demonstrated a successful cell-trapping and arraying. We believe this open microfluidics can be readily applied to other fluid channel designs.
3. Future Development
In the future, we are planning to further develop this system to construct the cell screening system combined with image AI recognition of cells. Furthermore, we will work on the application of bioproduction (ex. production of biological chemical products materials) with eyes on SDGs.
●The research was conducted with the support of the following research funds (Main Administration, Funding Agency, etc.)
Grants-in Aid for Scientific Research (Ministry of Education, Culture, Sports, Science and Technology-Japan).
Main research funds: Basic Study (S) (Principle Investigator: Hiroshi SHIMIZU, 19H05626) “High performance microbial cell factories development by model based metabolic design and adaptive laboratory”.
Partial research funds: Basic Study (B) (Principle Investigator: Hiroaki SUZUKI, 19H02576) “A Production Device for Lipid Bilayer Bioreactor and Its Application for Biomarker Detection Technology” and Grant-in-Aid for Transformative Research Areas (A) (Principal Investigator: Masahiro TAKINOUE, 20H05935) “DNA Nanoscale Modality”.
In addition to the above, this research was supported by JKA and its promotion funds from KEIRIN RACE.
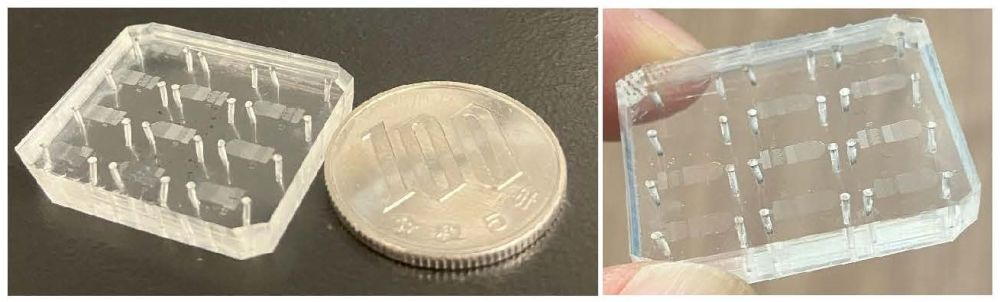
Figure 1. The actual open microfluidics made of silicone rubber. A large number of cell-trapping is realized on the small regions on the chip.
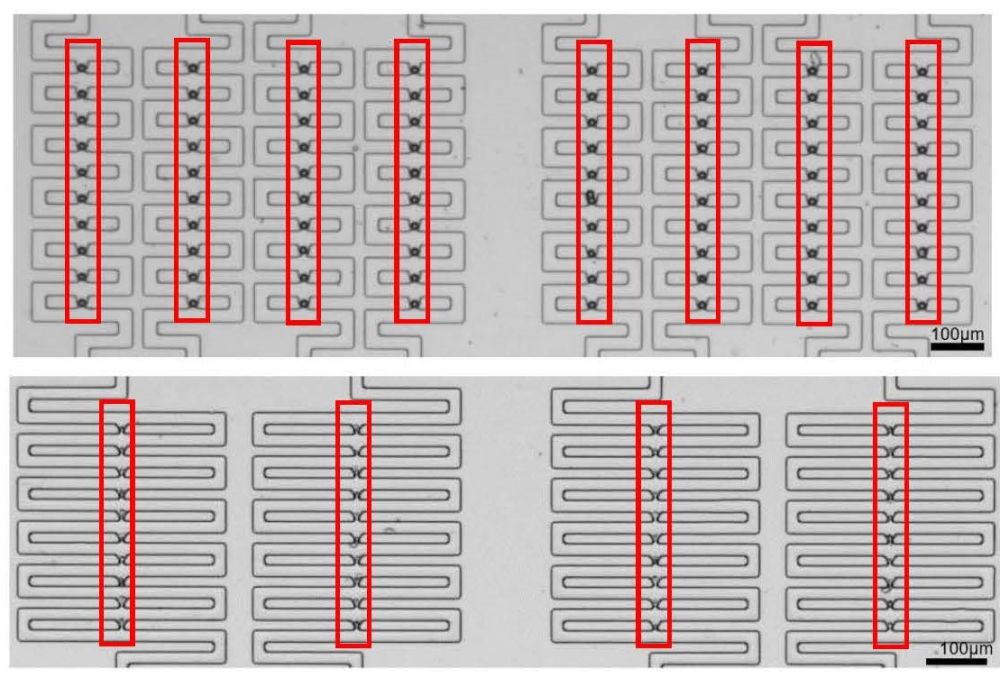
Figure 2. The trapping and an arraying of microbeads (upper image) and white blood cells (lowerimage) in the open microfluidics.
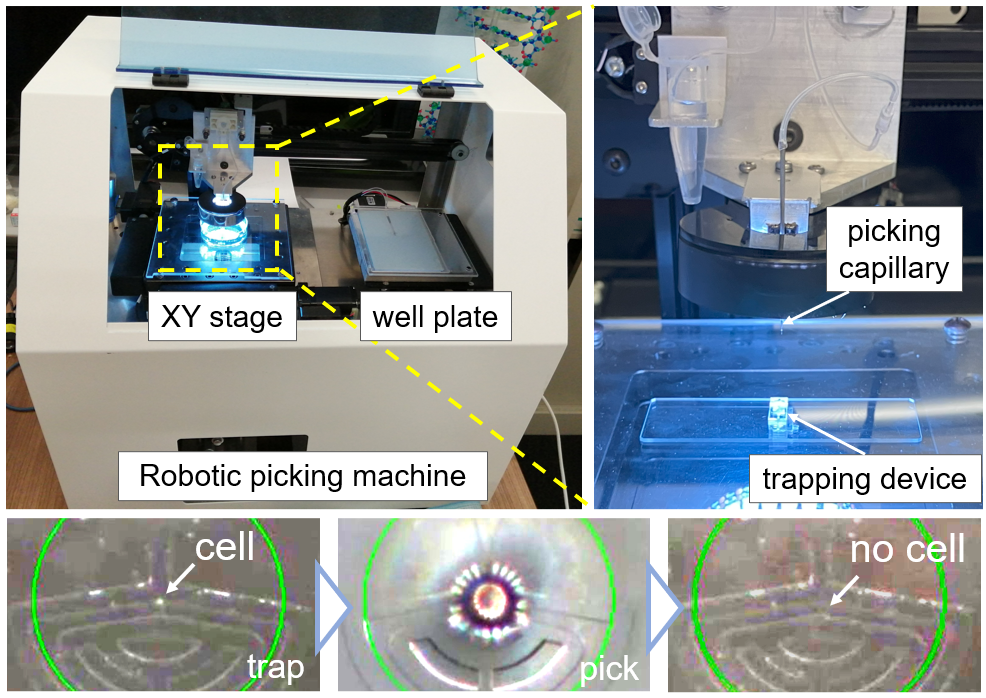
Figure 3. A robotic picking system using a glass capillary after trapping cells on the chip. The lower consecutive images show how the trapped cell is aspirated.
【Contact Information】
<About research>
Hiroaki SUZUKI
Professor, Faculty of Science and Engineering. Chuo University (Department of Precision Mechanics)
TEL: 03-3817-1827
E-mail: suzuki★mech.chuo-u.ac.jp
● Please replace ★ with @ when sending email.
<About Public Relations>
Research Support Office, Chuo University
TEL: 03-3817-7423 or 1675 FAX: 03-3817-1677
E-mail: kkouhou-grp★g.chuo-u.ac.jp
● Please replace ★ with @ when sending email.
【Glossary】
*1 Open microfluidics
In conventional microfluidics, the chip with a flow channel is bonded with another substrate to form a closed microchannel. Thus, it is common to apply pressure to drive the flow in these closed microfluidics. On the other hand, in open microfluidics, one side or both sides of the flow channel are open to the air so that they allow access to the flow channel from outside. Therefore, wetting phenomenon (spontaneous capillary flow) is employed to drive the flow instead of using the pressure pump.
*2 Single cell analysis and manipulation
A class of technologies that manipulate and analyze individual cells by separating them one by one instead of treating cells in the bulk. Microfluidics enables fluid control within the space in the size similar to that of cells, offering a high affinity with single-cell technology. By examining the features of individual cells, we expect to add a new dimension to biology and medical fields.
*3 Screening Technology
A technology to select and pick up specific (target) cells within the cell population. We expect it to contribute to facilitating bio-drug discovery and production of bio-chemical products through acquiring cells that highly produce useful substances, for instance.


